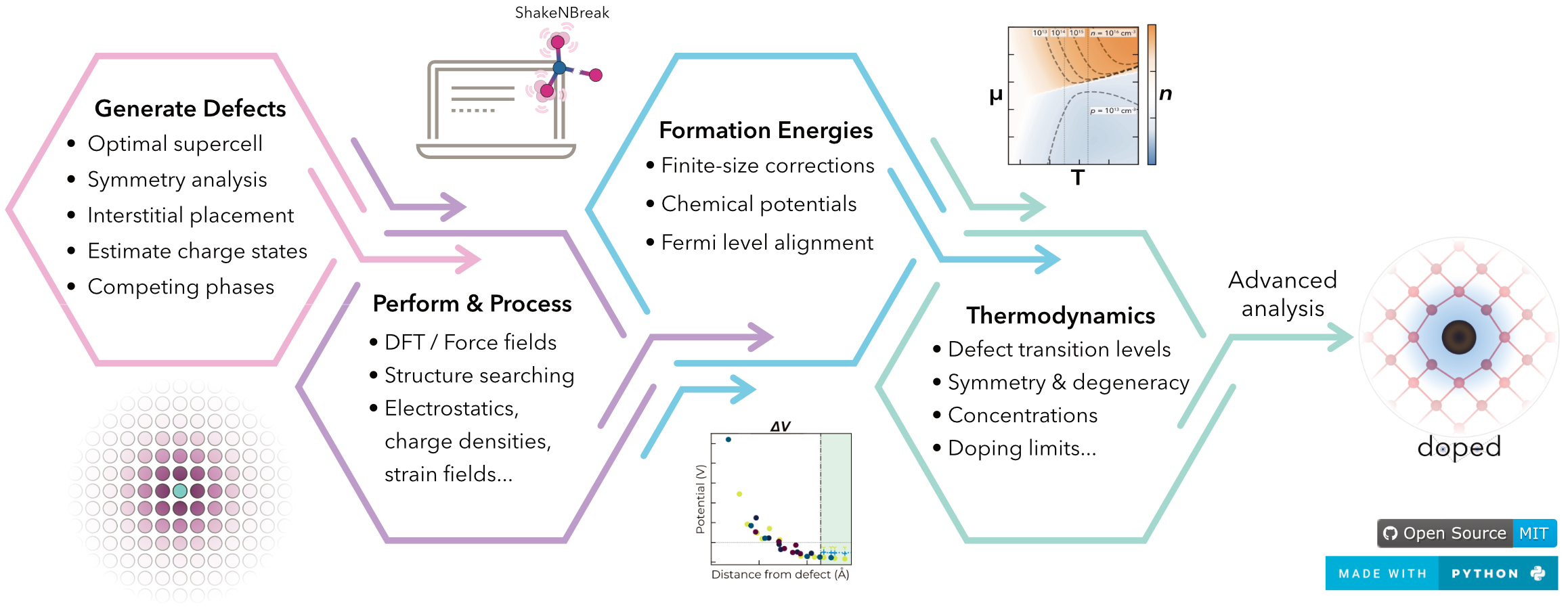
Codes
We lead and contribute to the development of a range of computational packages to support our research, primarily based in Python. These computational tools are open-source and are used by thousands of other researchers worldwide. Code development is not a requirement in our group, but it can be a great way to build and showcase highly-desired skills, while greatly accelerating our research and benefitting the global research community.
We aim to produce code that is simultaneously powerful and user-friendly, to maximise impact. These include:
doped
doped is a comprehensive Python suite for the generation and analysis of defect calculations, implementing the defect simulation workflow in an efficient, reproducible, user-friendly yet powerful and fully-customisable manner.
Click any of the images here to check out the docs sites!
ShakeNBreak
ShakeNBreak is a defect structure-searching method (i.e. global optimisation strategy) employing chemically-guided bond distortions to locate ground-state and metastable structures of defects in solid materials.
NequIP & Allegro

NequIP is an open-source code for building equivariant graph neural network (GNN) interatomic potentials (i.e. machine-learned force fields; MLFFs), which has inspired several other community codes in this area. See the matbench-discovery leaderboard for inorganic material foundation models.
Allegro is an extension to NequIP, which implements a strictly-local GNN architecture to facilitate simulations of millions of atoms 🚀
PyTASER
PyTASER is a Python library for simulating differential absorption spectra in compounds using quantum chemical calculations, including transient (TAS) and differential (DAS) absorption spectroscopies.
easyunfold
easyunfold implements electronic band structure unfolding, which is a technique for obtaining the effective band structure of symmetry-broken systems, such as disordered / defective / alloyed / thermally-active materials.
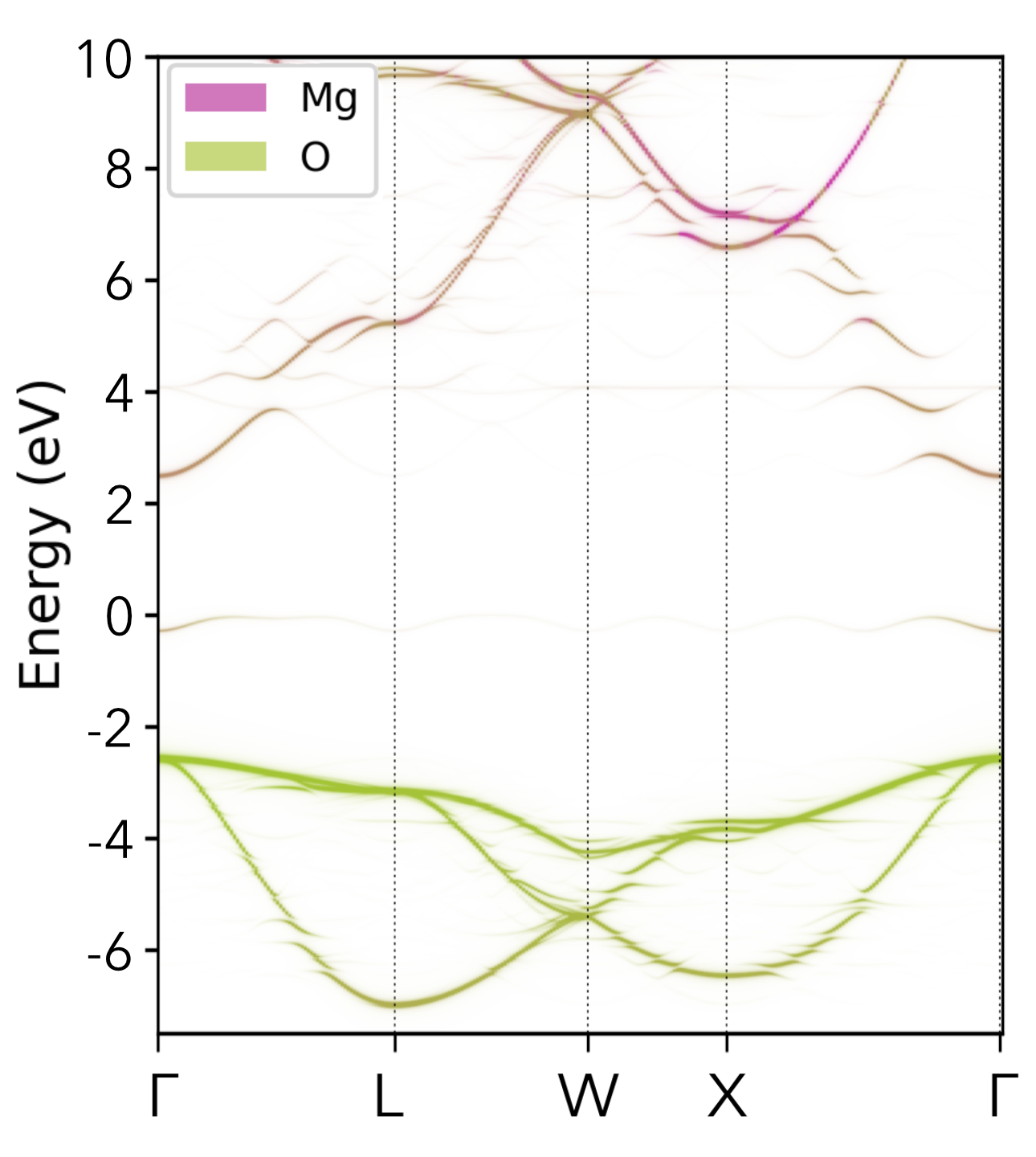
| Cs₂(Sn/Ti)Br₆ Vacancy-Ordered Perovskite Alloys | Oxygen Vacancy (Vₒ⁰) in MgO |
|---|---|
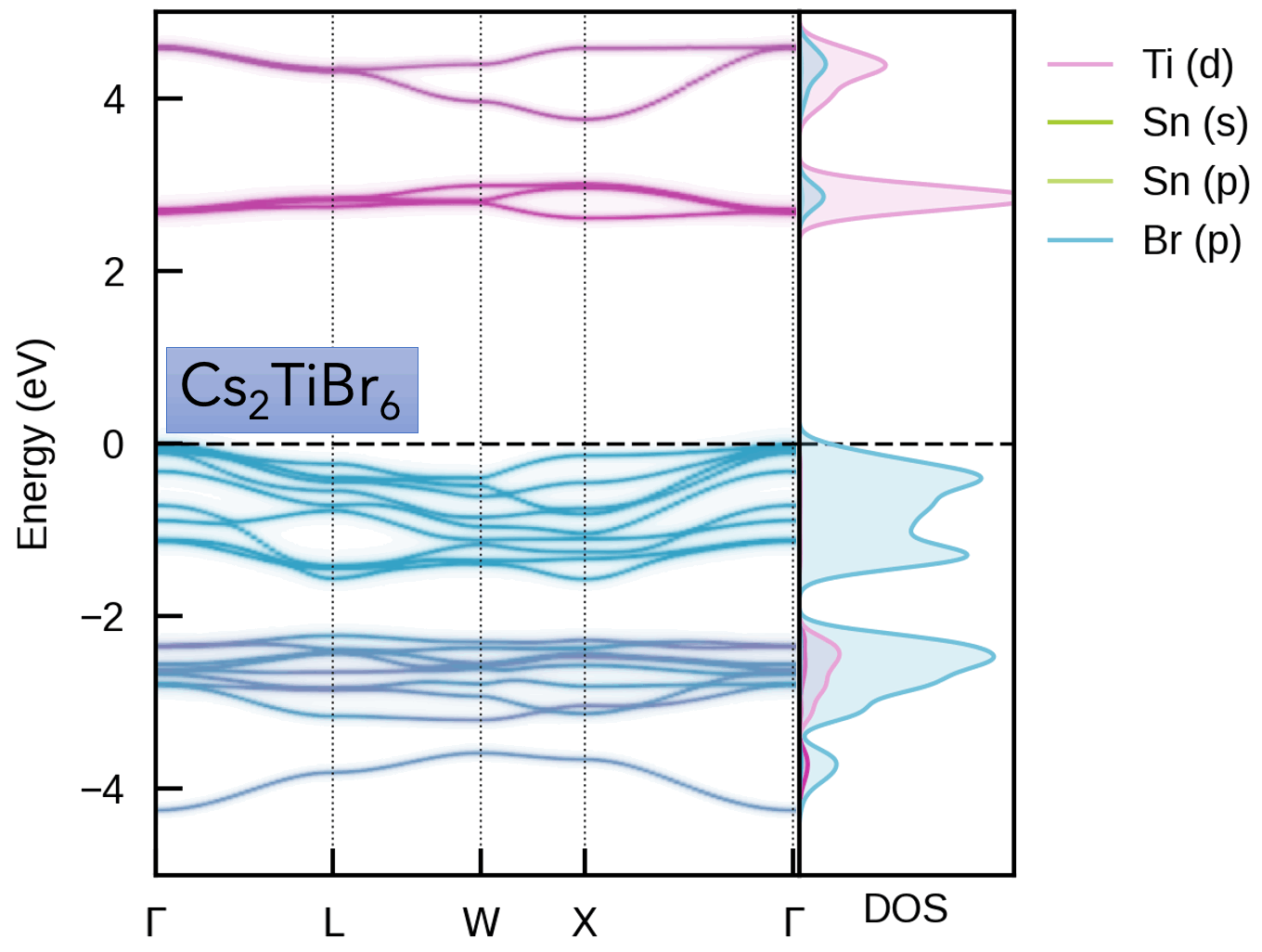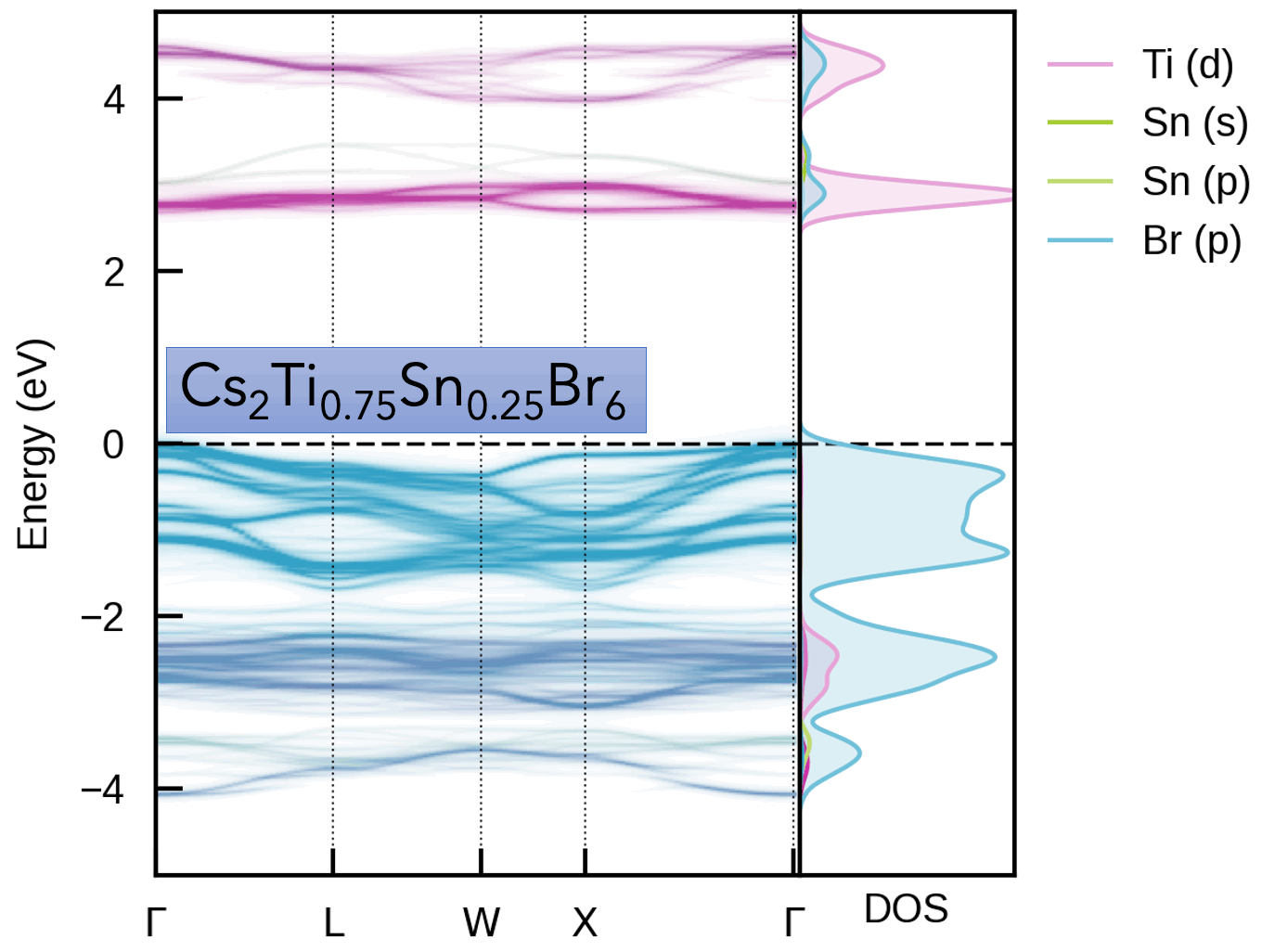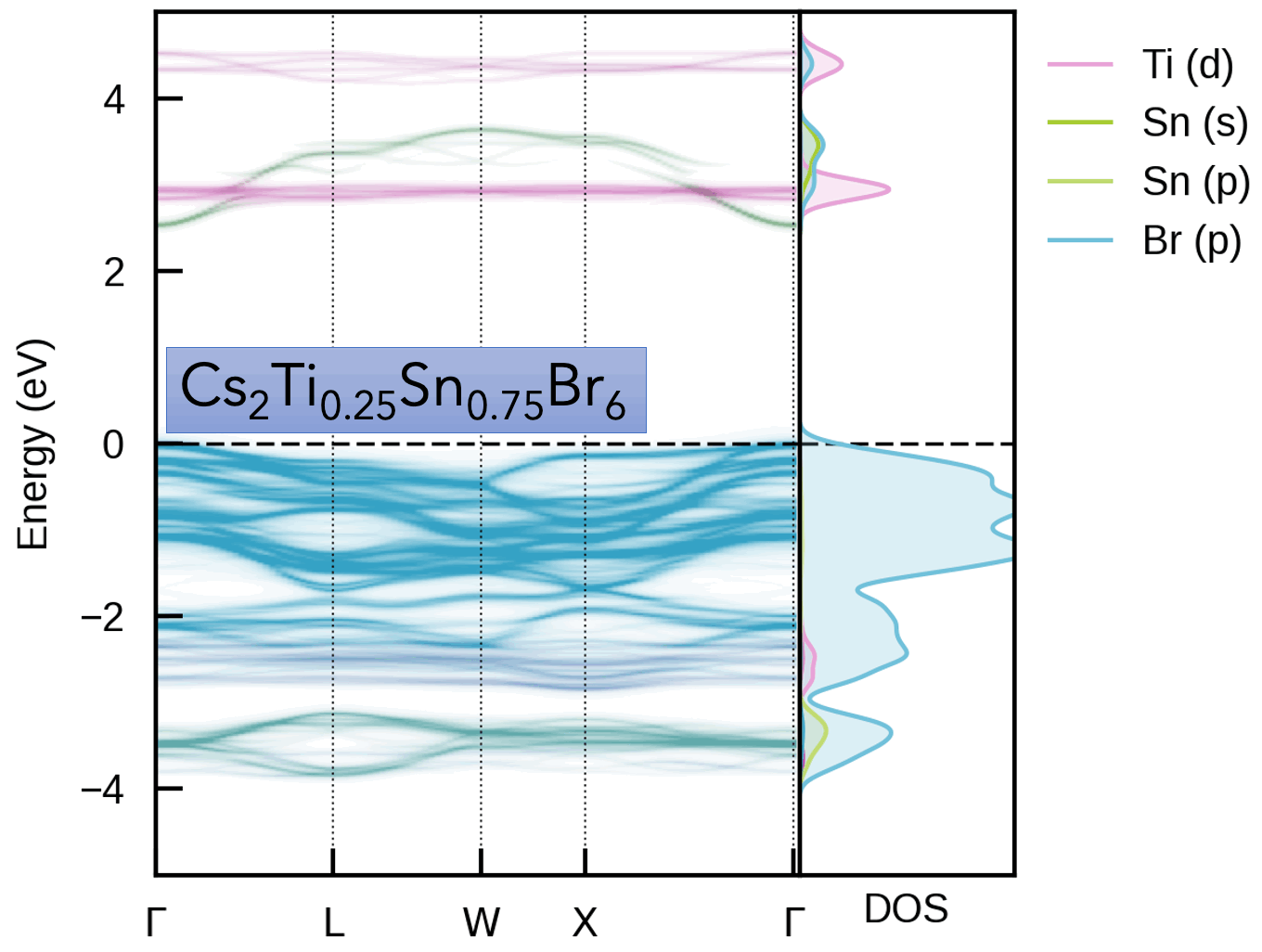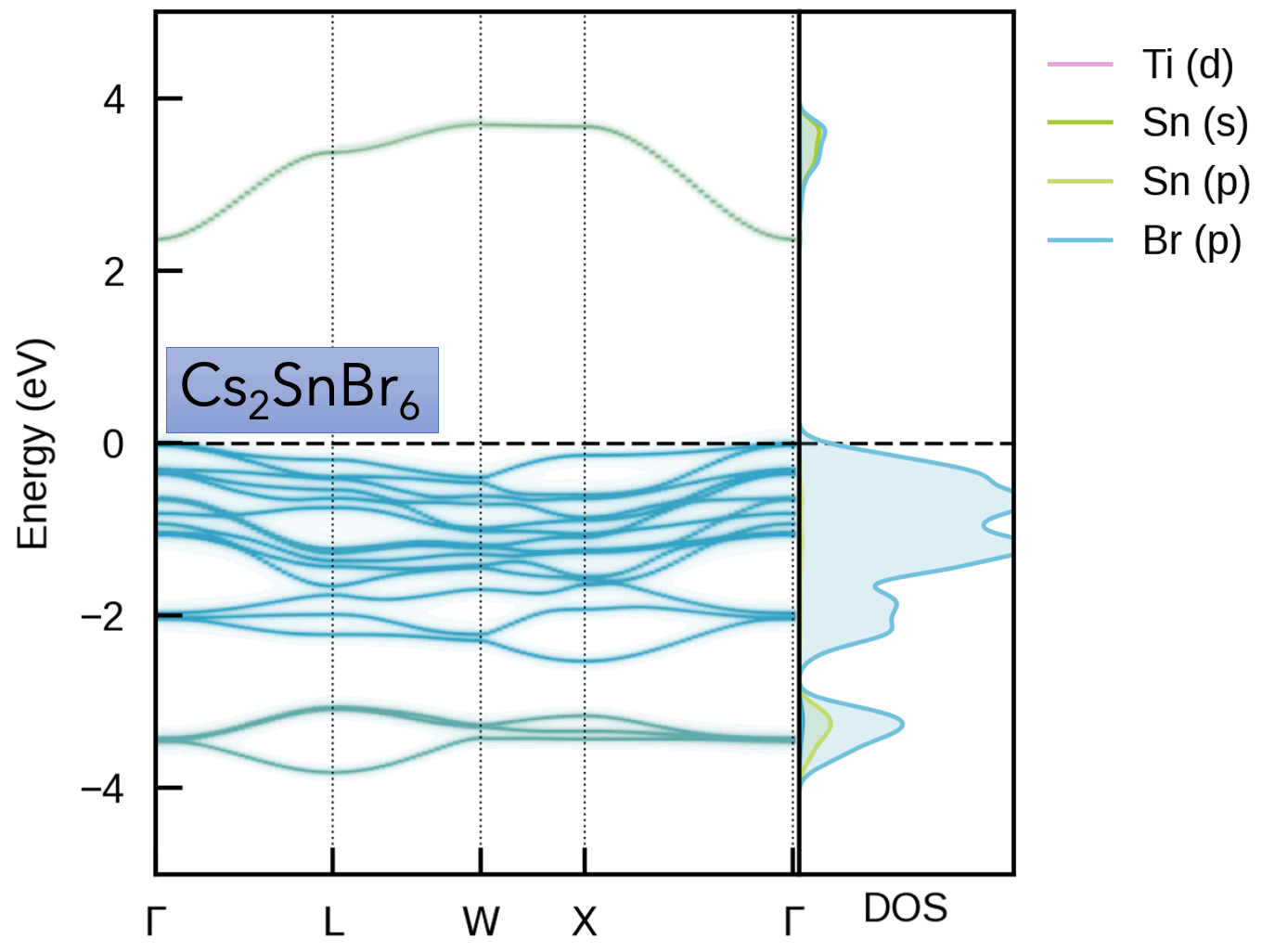 |  |
We also contribute to a number of community codes, including: